Discovery of Biomarkers for Endometrial Carcinoma
From LEAP
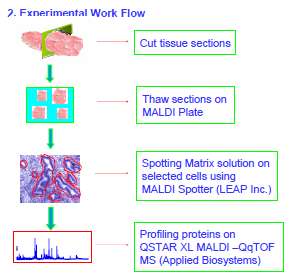
Purpose: Discovering biomarkers for endometrial carcinoma rapidly by direct profiling of proteins in tissue sections.
Methods: nano-scale MALDI-Spotting and MALDI QqTOF mass spectrometry.
Results: Five up-regulated protein markers for endometrial carcinoma were discovered with the new tool.
Discovery of Biomarkers for Endometrial Carcinoma by Direct Profiling of Proteins in Tissue Sections Using Selective MALDI-SPOTTING and MALDI MS
Jingzhong Guo (1); Muntajib Alhaq (2); Leroi V. DeSouza (1); Gordon Nye (3); Werner Martin (3); Alexander D. Romaschin (4); Terence J. Colgan (2); K..W. Michael Siu (1) 1 York University, Toronto, ON, Canada; 2 Mount Sinai Hospital, Toronto, ON, Canada; 3 LEAP Technologies, Inc., Carrboro, NC; 4 Toronto General Hospital, Toronto, ON, Canada
Introduction In recent years, various technologies have been used to acquire protein profiles (or maps) for biomarker discovery. Many technologies (e.g. 2D PAGE etc.) require multiple steps of sample handling and separation. These steps are time consuming and may lead to protein losses. Direct profiling of proteins in tissue sections provides an alternative and rapid way for biomarker discovery. The challenge is to acquire protein profiles from a user-defined group of cells (e.g. cancer). Here we report our recent work in discovering biomarkers for endometrial carcinoma by direct profiling proteins in tissue sections using a nano-scale MALDI spotter and MS.
Continued in Poster
Reprint of Poster
![]() ASMS 2005 Poster - Discovery of Biomarkers for Endometrial Carcinoma by Direct Profiling of Proteins in Tissue Sections Using Selective MALDI-Spotting and MALDI MS
ASMS 2005 Poster - Discovery of Biomarkers for Endometrial Carcinoma by Direct Profiling of Proteins in Tissue Sections Using Selective MALDI-Spotting and MALDI MS
Contact LEAP
For additional information about this technique please contact LEAP Technologies for detailed information